5. Vectorization#
This chapter introduces vectorization, a technique for encoding qualitative data (like words) into numeric values. We will use a data structure, the document-term matrix, to work with vectorized texts, discuss weighting strategies for managing high-frequency tokens, and train a classification model to distinguish style.
Data: 20 Henry James novels, collected by Jonathan Reeve and broken into chapters with Reeve’s chapterization tool. Labels are from David L. Hoover’s clustering of James’s novels
Credits: Portions of this chapter are adapted from the UC Davis DataLab’s Natural Language Processing for Data Science
5.1. Preliminaries#
We will need the following libraries:
import numpy as np
import pandas as pd
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import classification_report
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import Normalizer
from sklearn.decomposition import PCA
import seaborn as sns
import matplotlib.pyplot as plt
Corpus documents are stored in a DataFrame alongside other metadata.
corpus = pd.read_parquet("data/datasets/james_chapters.parquet")
print(corpus.info())
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 563 entries, 0 to 562
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 novel 563 non-null object
1 year 563 non-null int64
2 directory 563 non-null object
3 file 563 non-null object
4 chapter 563 non-null int64
5 style 563 non-null int64
6 hoover 563 non-null int64
7 tokens 563 non-null object
8 masked 563 non-null object
dtypes: int64(4), object(5)
memory usage: 39.7+ KB
None
Novels are divided into their component chapters. Use .groupby() to count how
many chapters there are for each novel.
grouped = corpus.groupby("novel")
chapters_per_novel = grouped["chapter"].count()
chapters_per_novel.to_frame(name = "chapters")
| chapters | |
|---|---|
| novel | |
| Ambassadors | 36 |
| Awkward Age | 32 |
| Bostonians | 42 |
| Confidence | 30 |
| Golden Bowl | 16 |
| Ivory Tower | 13 |
| Outcry | 20 |
| Portrait Of A Lady | 55 |
| Princess Casamassima | 49 |
| Reverberator | 15 |
| Roderick Hudson | 13 |
| Sacred Found | 14 |
| Spoils Poynton | 22 |
| The American | 30 |
| The Europeans | 12 |
| Tragic Muse | 51 |
| Washington Square | 35 |
| Watch And Ward | 11 |
| What Maisie Knew | 31 |
| Wings Of The Dove | 36 |
The style and hoover columns contain labels. The first demarcates early
James from late with the publication of What Maisie Knew in 1897.
style_counts = grouped[["year", "style"]].value_counts()
style_counts = style_counts.to_frame(name = "chapters")
style_counts.sort_values("style")
| chapters | |||
|---|---|---|---|
| novel | year | style | |
| Reverberator | 1888 | 0 | 15 |
| Watch And Ward | 1871 | 0 | 11 |
| Bostonians | 1886 | 0 | 42 |
| Confidence | 1879 | 0 | 30 |
| Washington Square | 1880 | 0 | 35 |
| Tragic Muse | 1890 | 0 | 51 |
| The Europeans | 1878 | 0 | 12 |
| Portrait Of A Lady | 1881 | 0 | 55 |
| Princess Casamassima | 1886 | 0 | 49 |
| The American | 1877 | 0 | 30 |
| Roderick Hudson | 1875 | 0 | 13 |
| Spoils Poynton | 1897 | 1 | 22 |
| Ambassadors | 1903 | 1 | 36 |
| What Maisie Knew | 1897 | 1 | 31 |
| Outcry | 1911 | 1 | 20 |
| Ivory Tower | 1917 | 1 | 13 |
| Golden Bowl | 1904 | 1 | 16 |
| Awkward Age | 1899 | 1 | 32 |
| Sacred Found | 1901 | 1 | 14 |
| Wings Of The Dove | 1902 | 1 | 36 |
The second uses Hoover’s grouping of James’s novels into four distinct phases.
hoover_counts = grouped[["year", "hoover"]].value_counts()
hoover_counts = hoover_counts.to_frame(name = "chapters")
hoover_counts.sort_values("hoover")
| chapters | |||
|---|---|---|---|
| novel | year | hoover | |
| Confidence | 1879 | 0 | 30 |
| Watch And Ward | 1871 | 0 | 11 |
| Washington Square | 1880 | 0 | 35 |
| Portrait Of A Lady | 1881 | 0 | 55 |
| Roderick Hudson | 1875 | 0 | 13 |
| The Europeans | 1878 | 0 | 12 |
| The American | 1877 | 0 | 30 |
| Reverberator | 1888 | 1 | 15 |
| Bostonians | 1886 | 1 | 42 |
| Tragic Muse | 1890 | 1 | 51 |
| Princess Casamassima | 1886 | 1 | 49 |
| Awkward Age | 1899 | 2 | 32 |
| What Maisie Knew | 1897 | 2 | 31 |
| Spoils Poynton | 1897 | 2 | 22 |
| Ambassadors | 1903 | 3 | 36 |
| Outcry | 1911 | 3 | 20 |
| Ivory Tower | 1917 | 3 | 13 |
| Golden Bowl | 1904 | 3 | 16 |
| Sacred Found | 1901 | 3 | 14 |
| Wings Of The Dove | 1902 | 3 | 36 |
Tokens for each chapter are stored as strings in tokens and masked.
Chapters have been tokenized with nltk.word_tokenize(). Why masked? That
column has had its proper noun tokens masked out with PN. You will see why
later on.
See masking code
def mask_proper_nouns(string, mask = "PN"):
"""Mask proper nouns in a string.
Parameters
----------
string : str
String to mask
mask : str
Masking label
Returns
-------
masked : str
The masked string
"""
# First, split the string into tokens
tokens = string.split()
# Then assign part-of-speech tags to those tokens. The output of this
# tagger is a list of tuples, where the first element is the token and the
# second is the tag
tagged = nltk.pos_tag(tokens)
# Create a new list to hold the output
masked = []
for (token, tag) in tagged:
# If the tag is "NNP", replace it with our mask
token = mask if tag == "NNP" else token
# Add the token to the output list
masked.append(token)
# Join the list and return
masked = " ".join(masked)
return masked
5.2. The Document-Term Matrix#
So far we have worked with lists of tokens. That works for some tasks, but to compare documents with one another, it would be better to represent our corpus as a two-dimensional array, or matrix. In this matrix, each row is a document and each column is a token; cells record the number of times that token appears in a document. The resultant matrix is known as the document-term matrix, or DTM.
It isn’t difficult to convert lists of tokens into a DTM, but scikit-learn
can do it. Unless you have a reason to convert your token lists manually, just
rely on that.
See function to create a document-term matrix by hand
def make_dtm(docs):
"""Make a document-term matrix.
Parameters
----------
docs : list[str]
A list of strings
Returns
-------
dtm, vocabulary : tuple[list, set]
The document-term matrix and the corpus vocabulary
"""
# Split the documents into tokens
docs = [doc.split() for doc in docs]
# Get the unique set of tokens for all documents in the corpus
vocab = set()
for doc in docs:
# A set union (`|=`) adds any new tokens from the current document to
# the running set of all tokens
vocab |= set(doc)
# Create a list of m dictionaries, where m is the number of corpus
# documents. Each dictionary will have every token in the vocabulary (key),
# which is initially assigned to 0 (value)
counts = [dict.fromkeys(vocab, 0) for doc in docs]
# Roll through each document
for idx, doc in enumerate(docs):
# For each token in a document...
for tok in doc:
# Access the document counts, then access the token stored in the
# dictionary. Increment the corresponding count by 1
counts[idx][tok] += 1
# Extract the values from each dictionary
dtm = [[count for count in doc.values()] for doc in counts]
# Return the DTM and the vocabulary
return dtm, vocab
Many classes in scikit-learn have the same use pattern: first, you initialize
the class by assigning it to a variable (and optionally set parameters), then
you fit it on your data. The CountVectorizer, which makes a DTM, does
just this. It will even tokenize strings while it fits, though watch out: it
has a simple tokenization pattern, so it’s often best to do this step yourself.
Below, we initialize the CountVectorizer and set the following parameters:
token_pattern: a regex pattern for which tokens to keep (here: any alphabetic characters of three or more characters)stop_words: remove function words in Englishstrip_accents: normalize accents to ASCII
cv_parameters = {
"token_pattern": r"\b[a-zA-Z]{3,}\b",
"stop_words": "english",
"strip_accents": "ascii"
}
count_vectorizer = CountVectorizer(**cv_parameters)
count_vectorizer.fit(corpus["tokens"])
CountVectorizer(stop_words='english', strip_accents='ascii',
token_pattern='\\b[a-zA-Z]{3,}\\b')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
CountVectorizer(stop_words='english', strip_accents='ascii',
token_pattern='\\b[a-zA-Z]{3,}\\b')With the vectorizer fitted, transform the data you fitted it on.
dtm = count_vectorizer.transform(corpus["tokens"])
DTMs are sparse. That is, they are mostly made up of zeros.
dtm
<Compressed Sparse Row sparse matrix of dtype 'int64'
with 504302 stored elements and shape (563, 27822)>
This sparsity is significant. Comparing documents with each other requires taking into account all unique tokens in the corpus, not just those in a particular document. This means we must count the number of times a token appears in a document even if that count is zero. What those zero counts also mean is that the documents in a DTM are not strictly those documents that are in the corpus. They are potential texts: possible distributions of tokens across the corpus.
The output of CountVectorizer is optimized for keeping the memory footprint
of a DTM low. But for a small corpus like this, use .toarray() to convert the
matrix into a NumPy array.
dtm = dtm.toarray()
Now, wrap this as a DataFrame and set the column names with the output of the
vectorizer’s .get_feature_names_out() method.
dtm = pd.DataFrame(
dtm, columns = count_vectorizer.get_feature_names_out()
)
dtm.head()
| aback | abandon | abandoned | abandoning | abandonment | abandons | abase | abased | abasement | abash | ... | zero | zest | zigzags | zola | zone | zones | zoo | zoological | zouaves | zurich | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 27822 columns
This DTM is indexed in the same order as the corpus documents. But for
readability’s sake, set the index to the novel and chapter columns of our
corpus DataFrame. Be sure to change the .names attribute of the index, or
your index will conflict with possible column values in the DTM.
dtm.index = pd.MultiIndex.from_arrays(
[corpus["novel"], corpus["chapter"]],
names = ["novel_name", "chapter_num"]
)
dtm.head()
| aback | abandon | abandoned | abandoning | abandonment | abandons | abase | abased | abasement | abash | ... | zero | zest | zigzags | zola | zone | zones | zoo | zoological | zouaves | zurich | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| novel_name | chapter_num | |||||||||||||||||||||
| Watch And Ward | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 27822 columns
5.2.1. Document-term matrix analysis#
Numeric operations across the DTM now work the same as they would for any other DataFrame. Here, total tokens per novel:
chapter_token_count = np.sum(dtm, axis = 1).to_frame(name = "token_count")
chapter_token_count.groupby("novel_name").sum()
| token_count | |
|---|---|
| novel_name | |
| Ambassadors | 56551 |
| Awkward Age | 48957 |
| Bostonians | 63102 |
| Confidence | 29888 |
| Golden Bowl | 74098 |
| Ivory Tower | 23593 |
| Outcry | 21398 |
| Portrait Of A Lady | 86844 |
| Princess Casamassima | 79966 |
| Reverberator | 21043 |
| Roderick Hudson | 54094 |
| Sacred Found | 25920 |
| Spoils Poynton | 26696 |
| The American | 53494 |
| The Europeans | 24071 |
| Tragic Muse | 81287 |
| Washington Square | 24444 |
| Watch And Ward | 25583 |
| What Maisie Knew | 36369 |
| Wings Of The Dove | 66442 |
On average, which three chapters are the longest across all of James’s novels?
chapter_avg_tokens = chapter_token_count.groupby("chapter_num").mean()
chapter_avg_tokens.sort_values("token_count", ascending = False).head(3)
| token_count | |
|---|---|
| chapter_num | |
| 5 | 2237.000000 |
| 46 | 2223.666667 |
| 14 | 2081.687500 |
What is the average chapter length?
chapter_avg_tokens.mean()
token_count 1610.572704
dtype: float64
Top ten chapters with the highest type counts:
num_types = (dtm > 0).sum(axis = 1).to_frame(name = "num_types")
num_types.nlargest(10, "num_types")
| num_types | ||
|---|---|---|
| novel_name | chapter_num | |
| Golden Bowl | 14 | 3991 |
| 5 | 3990 | |
| 9 | 3565 | |
| 11 | 3129 | |
| Roderick Hudson | 3 | 2468 |
| 1 | 2242 | |
| 11 | 2242 | |
| Golden Bowl | 7 | 2240 |
| Roderick Hudson | 10 | 2174 |
| Golden Bowl | 10 | 2085 |
Most frequent word in each novel:
token_freq = dtm.groupby("novel_name").sum()
token_freq.idxmax(axis = 1).to_frame(name = "most_frequent_token")
| most_frequent_token | |
|---|---|
| novel_name | |
| Ambassadors | strether |
| Awkward Age | mrs |
| Bostonians | verena |
| Confidence | bernard |
| Golden Bowl | maggie |
| Ivory Tower | gray |
| Outcry | lord |
| Portrait Of A Lady | isabel |
| Princess Casamassima | hyacinth |
| Reverberator | francie |
| Roderick Hudson | rowland |
| Sacred Found | mrs |
| Spoils Poynton | fleda |
| The American | newman |
| The Europeans | said |
| Tragic Muse | nick |
| Washington Square | catherine |
| Watch And Ward | roger |
| What Maisie Knew | mrs |
| Wings Of The Dove | kate |
All are proper nouns, which often happens with fiction. In one way, this is valuable information: if you were modeling a corpus with different kinds of documents, you might use names’ frequency to distinguish fiction. But we only have James’s novels, and the high frequency of names can make it difficult to identify similarities across documents.
5.2.2. Using masked tokens#
This is where the text stored in masked comes in. That text masks over proper
nouns and treats them all like the same token. More, due to the way we’ve
currently configured our DTM generation, those masks will be dropped because
they’re only two characters long. That’s perfectly fine for our purposes. But
it again underscores the fact that documents in the DTM are not documents as
they are in the corpus. Indeed, through these preprocessing decisions we have
already constructed a model of our corpus.
Time to rebuild the DTM with text in masked.
count_vectorizer = CountVectorizer(**cv_parameters)
count_vectorizer.fit(corpus["masked"])
dtm = count_vectorizer.transform(corpus["masked"])
Convert to a DataFrame.
dtm = pd.DataFrame(
dtm.toarray(), columns = count_vectorizer.get_feature_names_out()
)
dtm.index = pd.MultiIndex.from_arrays(
[corpus["novel"], corpus["chapter"]],
names = ["novel_name", "chapter_num"]
)
We won’t step through the above metrics again, except we will look at top token counts to confirm that masking made a difference.
token_freq = dtm.groupby("novel_name").sum()
token_freq.idxmax(axis = 1).to_frame(name = "most_frequent_token")
| most_frequent_token | |
|---|---|
| novel_name | |
| Ambassadors | little |
| Awkward Age | know |
| Bostonians | said |
| Confidence | said |
| Golden Bowl | little |
| Ivory Tower | said |
| Outcry | said |
| Portrait Of A Lady | said |
| Princess Casamassima | said |
| Reverberator | said |
| Roderick Hudson | said |
| Sacred Found | little |
| Spoils Poynton | said |
| The American | said |
| The Europeans | said |
| Tragic Muse | said |
| Washington Square | said |
| Watch And Ward | said |
| What Maisie Knew | little |
| Wings Of The Dove | said |
Names are gone but the output looks even worse. There’s no differentiation among the most frequent tokens in each novel, even when controlling for common deictic words with stopword removal. Given the nature of Zipfian distributions, this shouldn’t be surprising.
One way to control for this would be to remove tokens from consideration when building the DTM using some cutoff metric. That would work okay but it may remove valuable information from the documents. Consider, for example, the fact that James’s penchant for extended psychological descriptions could be usefully counterposed with chapters with more dialogue. Removing “said” would make it difficult to do this. More, setting the cutoff point could take a fair bit of back and forth. A better strategy would be to re-weight token counts by some method so that frequent tokens have less impact in aggregate analyses like the above.
5.3. Weighting with TF–IDF#
This is where TF–IDF, or term frequency–inverse document frequency, comes in. It re-weights tokens according to their specificity in a document. Tokens that frequently appear in many documents will have low TF–IDF scores, while those that are less frequent, or appear frequently in only a few documents, will have high TF–IDF scores.
Scores are the product of two statistics: term frequency and inverse document frequency. There are several variations for calculating both but generally they work like so:
Term frequency is the relative frequency of a token \(t\) in a document \(d\).
Where:
\(f_{t,d}\) is the frequency of token \(t\) in document \(d\)
\(\sum_{i=1}^nf_{i,d}\) is the sum of all token frequencies in document \(d\)
In code, that looks like the following:
TF = dtm.div(dtm.sum(axis = 1), axis = 0)
Inverse document frequency measures how common or rare a token is.
Where:
\(N\) is the total number of documents in a corpus \(D\)
For each document \(d\) in \(D\), we count which ones contain token \(t\)
The code for this calculation is below. Note that we typically add one to the document frequency to avoid zero-division errors. Adding one outside the logarithm ensures that any terms that appear across all documents do not completely zero-out.
N = len(dtm)
DF = (dtm > 0).sum(axis = 0)
IDF = np.log(1 + N / (1 + DF)) + 1
The product of these two statistics is TF–IDF.
Or, in code:
TFIDF = TF.multiply(IDF, axis = 1)
Don’t want to go through all those steps? Use TfidfVectorizer. But note that
scikit-learn has set some defaults for smoothing/normalizing TF–IDF scores
that could make the result slightly different than your own calculations.
Fitting TfidfVectorizer works with the same use pattern.
tfidf_vectorizer = TfidfVectorizer(**cv_parameters)
tfidf_vectorizer.fit(corpus["masked"])
tfidf = tfidf_vectorizer.transform(corpus["masked"])
Convert to a DataFrame:
tfidf = pd.DataFrame(
tfidf.toarray(), columns = tfidf_vectorizer.get_feature_names_out()
)
tfidf.index = pd.MultiIndex.from_arrays(
[corpus["novel"], corpus["chapter"]],
names = ["novel_name", "chapter_num"]
)
And now, finally, the highest scoring tokens for every novel. Again, these are the most specific tokens.
max_tfidf_per_novel = tfidf.groupby("novel_name").max()
max_tfidf_per_novel.idxmax(axis = 1).to_frame(name = "top_token")
| top_token | |
|---|---|
| novel_name | |
| Ambassadors | little |
| Awkward Age | know |
| Bostonians | policeman |
| Confidence | vivian |
| Golden Bowl | bowl |
| Ivory Tower | uncle |
| Outcry | outcry |
| Portrait Of A Lady | dance |
| Princess Casamassima | fiddler |
| Reverberator | germain |
| Roderick Hudson | prince |
| Sacred Found | grow |
| Spoils Poynton | negotiation |
| The American | marquis |
| The Europeans | said |
| Tragic Muse | boat |
| Washington Square | father |
| Watch And Ward | cousin |
| What Maisie Knew | ladyship |
| Wings Of The Dove | marian |
Top-scoring tokens for each chapter in What Maisie Knew. Use an empty slice
to get all entries in the second of the two DataFrame indexes.
maisie = tfidf.loc[("What Maisie Knew", slice(None))]
maisie_max = pd.DataFrame({
"token": maisie.idxmax(axis = 1),
"tfidf": maisie.max(axis = 1)
})
maisie_max
| token | tfidf | |
|---|---|---|
| chapter_num | ||
| 1 | nurse | 0.186544 |
| 2 | lies | 0.204308 |
| 3 | papa | 0.285493 |
| 4 | diadem | 0.219282 |
| 5 | brougham | 0.173980 |
| 6 | governess | 0.239340 |
| 7 | papa | 0.196547 |
| 8 | papa | 0.238696 |
| 9 | ladyship | 0.364359 |
| 10 | mamma | 0.275737 |
| 11 | ladyship | 0.293165 |
| 12 | ladyship | 0.303566 |
| 13 | child | 0.153145 |
| 14 | child | 0.155951 |
| 15 | papa | 0.156633 |
| 16 | mother | 0.276220 |
| 17 | squared | 0.147945 |
| 18 | papa | 0.141638 |
| 19 | father | 0.161065 |
| 20 | ladyship | 0.194401 |
| 21 | ladyship | 0.209321 |
| 22 | fishwife | 0.134534 |
| 23 | ladyship | 0.143543 |
| 24 | afraid | 0.141128 |
| 25 | pays | 0.159513 |
| 26 | sands | 0.128149 |
| 27 | come | 0.149019 |
| 28 | divorce | 0.212882 |
| 29 | salon | 0.216161 |
| 30 | waiter | 0.223491 |
| 31 | pupil | 0.163950 |
5.4. Document Classification#
Each document in the weighted DTM is now a feature vector: a sequence of values that encode information about token distributions. These vectors allow us to estimate joint probabilities between features, which enables classification tasks.
5.4.1. The Multinomial Naive Bayes classifier#
We use a Multinomial Naive Bayes model to classify documents according to their assigned label, or class. The model trains by calculating the prior probability for each class. Then, it computes the posterior probability of each token given a class. The class with the highest posterior probability is selected as the label for a document.
Term |
Definition |
|---|---|
Naive Bayes |
Assumes conditionally independent features |
Multinomial distribution |
Models probabilities of counts across categories |
Prior probability |
Probability of an event before observing new data |
Posterior probability |
Probability of an event after observing new data |
Argmax (maximum likelihood estimation) |
Predicts class with highest posterior probability |
The formula for our classifier is as follows:
Where:
\(P(C_k)\): prior probability of class \(C_k\)
\(P(x_i|C_k)\): posterior probability of feature \(x_i\) given class \(C_k\)
\(P(C_k|x)\): probability of feature vector \(x\) being class \(C_k\)
5.4.2. Training a classifier#
No need to do this math ourselves; scikit-learn can do it. But first, we
split our data and their corresponding labels into training and testing
datasets. The model will train on the former, and we will validate that model
on the latter (which is data it hasn’t yet seen).
X_train, X_test, y_train, y_test = train_test_split(
tfidf, corpus["hoover"], test_size = 0.3, random_state = 357
)
print(f"Train set size: {len(X_train)}\nTest set size: {len(X_test)}")
Train set size: 394
Test set size: 169
Train the model using the same initialization/fitting pattern from before.
classifier = MultinomialNB(alpha = 0.005)
classifier.fit(X_train, y_train)
MultinomialNB(alpha=0.005)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
MultinomialNB(alpha=0.005)
5.4.3. Model diagnostics#
Use the .score() method to return the mean accuracy for all labels given test
data. This is the number of correct predictions divided by the total number of
true labels.
accuracy = classifier.score(X_test, y_test)
print(f"Model accuracy: {accuracy:.4f}%")
Model accuracy: 0.9645%
Generate a classification report to get a class-by-class summary of the classifier’s performance. This requires you to make predictions on the test size, which you then compare against the true labels.
preds = classifier.predict(X_test)
Now make and print the report.
periods = ["1871-81", "1886-90", "1896-99", "1901-17"]
report = classification_report(y_test, preds, target_names = periods)
print(report)
precision recall f1-score support
1871-81 1.00 1.00 1.00 58
1886-90 0.98 1.00 0.99 51
1896-99 1.00 0.81 0.89 26
1901-17 0.87 0.97 0.92 34
accuracy 0.96 169
macro avg 0.96 0.94 0.95 169
weighted avg 0.97 0.96 0.96 169
The above scores describe trade-offs between the following kinds of predictions:
Prediction |
Explanation |
Shorthand |
|---|---|---|
True positive |
Correctly predicts class of interest |
TP |
True negative |
Correctly predicts all other classes |
TN |
False positive |
Incorrectly predicts class of interest |
FP |
False negative |
Incorrectly predicts all other classes |
FN |
Here is a breakdown of score types:
Score |
Explanation |
Formula |
|---|---|---|
Precision |
Accuracy of positive predictions |
\(P = \frac{TP}{TP + FP}\) |
Recall |
Ability to find all relevant instances |
\(R = \frac{TP}{TP + FN}\) |
F1 |
A balance of precision and recall |
\(F1 = 2\times \frac{P\times R}{P + R}\) |
A weighted average of these scores offsets each class by its proportion in the testing set; the macro average reports scores with no weighting. The support for each class is the number of documents labeled with that class.
This model does extremely well. In fact, it may be a touch overfitted: too closely matched with its training data and therefore incapable of generalizing beyond that data. For our purposes this is less of a concern because the corpus and analysis are both constrained, but you might be suspicious of high-scoring results like this in other cases.
5.4.4. Top tokens per class#
Recall that the classifier makes its decisions based on the posterior probability of a feature vector. That means there are certain tokens in the corpus that are most likely to appear for each class. What are they?
First, extract the feature names, the class labels, and the log probabilities for each feature.
feature_names = tfidf_vectorizer.get_feature_names_out()
class_labels = classifier.classes_
log_probs = classifier.feature_log_prob_
Now iterate through every class and extract the top_n tokens (most probable
tokens).
top_n = 25
for idx, label in enumerate(class_labels):
# Get the probabilities and sort them. Sorting is in ascending order, so
# flip the array
probs = log_probs[idx]
sorted_probs = np.argsort(probs)[::-1]
# The above array contains the indexes that would sort the probabilities.
# Take the `top_n` indices, then get the corresponding tokens by indexing
# `feature_names`
top_probs = sorted_probs[:top_n]
top_tokens = feature_names[top_probs]
# Print the results
print(f"Top tokens for {label}:")
print("\n".join(top_tokens), end = "\n\n")
Top tokens for 0:
said
little
don
know
say
think
good
young
like
great
man
come
time
moment
father
asked
looked
make
girl
went
shall
looking
eyes
vivian
old
Top tokens for 1:
said
little
know
like
don
young
come
say
man
didn
time
good
think
great
way
make
moment
old
girl
people
want
went
lady
thought
mother
Top tokens for 2:
know
said
little
don
say
mother
time
child
just
like
come
way
quite
mean
think
friend
make
really
things
moment
dear
good
looked
old
thing
Top tokens for 3:
said
know
little
time
don
quite
come
moment
just
really
way
mean
say
like
friend
question
make
man
things
fact
didn
thing
good
think
want
These are pretty general. Even with tf-idf, common tokens in fiction persist. But we can compute the difference between log probabilities for one class and the mean log probabilities of all other classes. That will give us more distinct tokens for each class.
for idx, label in enumerate(class_labels):
# Remove the current classes's log probabilities, then calculate their mean
other_classes = np.delete(log_probs, idx, axis = 0)
mean_log_probs = np.mean(other_classes, axis = 0)
# Find the difference between this mean and the current class's log
# probabilities
difference = log_probs[idx] - mean_log_probs
# Sort as before
sorted_probs = np.argsort(difference)[::-1]
top_probs = sorted_probs[:top_n]
top_tokens = feature_names[top_probs]
# And print
print(f"Distinctive tokens for {label}:")
print("\n".join(top_tokens), end = "\n\n")
Distinctive tokens for 0:
vivian
osmond
marquis
honor
favor
recognized
humor
townsend
color
colored
parlor
catherine
monsieur
ardor
countess
evers
chateau
confectioner
neighbors
neighboring
isabel
misfortunes
dishonor
rowland
bruises
Distinctive tokens for 1:
burrage
agnes
bookbinder
univers
abbey
olive
fiddler
farrinder
proberts
embassy
dressmaker
millicent
poppa
tarrant
plebeian
canvases
electors
stile
verena
boulevard
mediocrity
intonations
heroes
comedian
daresay
Distinctive tokens for 2:
straighteners
farange
maltese
fleda
negotiation
diadem
stepfather
owen
rolls
avez
hug
banister
vanderbank
schoolroom
pelisse
pupil
curate
miser
maisie
wix
profitably
frightening
tishy
texts
overmore
Distinctive tokens for 3:
strether
crimble
milly
lowder
connections
chad
pococks
insistent
clearance
pounce
enhance
sagely
entresol
murmurous
psychologic
embroider
reaffirmed
intensified
clues
cessation
tortoise
densher
disclaimed
showily
hillside
5.5. Visualization#
Let’s visualize our documents in a scatterplot so we can inspect the corpus at scale.
5.5.1. Dimensionality reduction#
To do this, we’ll need to transform our TF–IDF vectors into simplified representations. Right now, these vectors are extremely high dimensional:
_, num_dimensions = tfidf.shape
print(f"Number of dimensions: {num_dimensions:,}")
Number of dimensions: 26,053
This number far exceeds the two or three dimensions of plots.
We use principal component analysis, or PCA, to reduce the dimensionality of our vectors so we can plot them. PCA identifies axes (principal components) that maximize variance in data and then projects that data onto the components. This reduces the number of features in the data but retains important information about each vector. Take a look at Margaret Fleck’s lecture notes if you’d like to see how this process works in detail.
pca = PCA(0.95, random_state = 357)
pca.fit(tfidf)
PCA(n_components=0.95, random_state=357)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
PCA(n_components=0.95, random_state=357)
The PCA reducer’s .explained_variance_ratio_ attribute contains the
proportion of the total variance captured by each principal component. Their
sum should equal the number we set above.
exp_variance = np.sum(pca.explained_variance_ratio_)
print(f"Explained variance: {exp_variance:.2f}%")
Explained variance: 0.95%
Slice out segments of these components to identify how much of the variance is
explained by the \(k\)-th component. scikit-learn sorts components
automatically, so the first ones always contain the most variance..
k = 25
exp_variance = np.sum(pca.explained_variance_ratio_[:k])
print(f"Explained variance of {k} components: {exp_variance:.2f}%")
Explained variance of 25 components: 0.15%
The first two components do not explain very much variance, but they will be enough for visualization.
k = 2
exp_variance = np.sum(pca.explained_variance_ratio_[:k])
print(f"Explained variance of {k} components: {exp_variance:.2f}%")
Explained variance of 2 components: 0.04%
5.5.2. Plotting documents#
To plot, transform the TF–IDF scores and format the reduced data as a DataFrame.
reduced = pca.transform(tfidf)
vis_data = pd.DataFrame(reduced[:, 0:2], columns = ["x", "y"])
vis_data["label_idx"] = corpus["hoover"].copy()
vis_data["label"] = vis_data["label_idx"].map(lambda x: periods[x])
Create a plot.
plt.figure(figsize = (10, 10))
g = sns.scatterplot(
x = "x",
y = "y",
hue = "label",
palette = "tab10",
alpha = 0.8,
data = vis_data,
)
g.set(title = "James chapters", xlabel = "Dim. 1", ylabel = "Dim. 2")
plt.show()
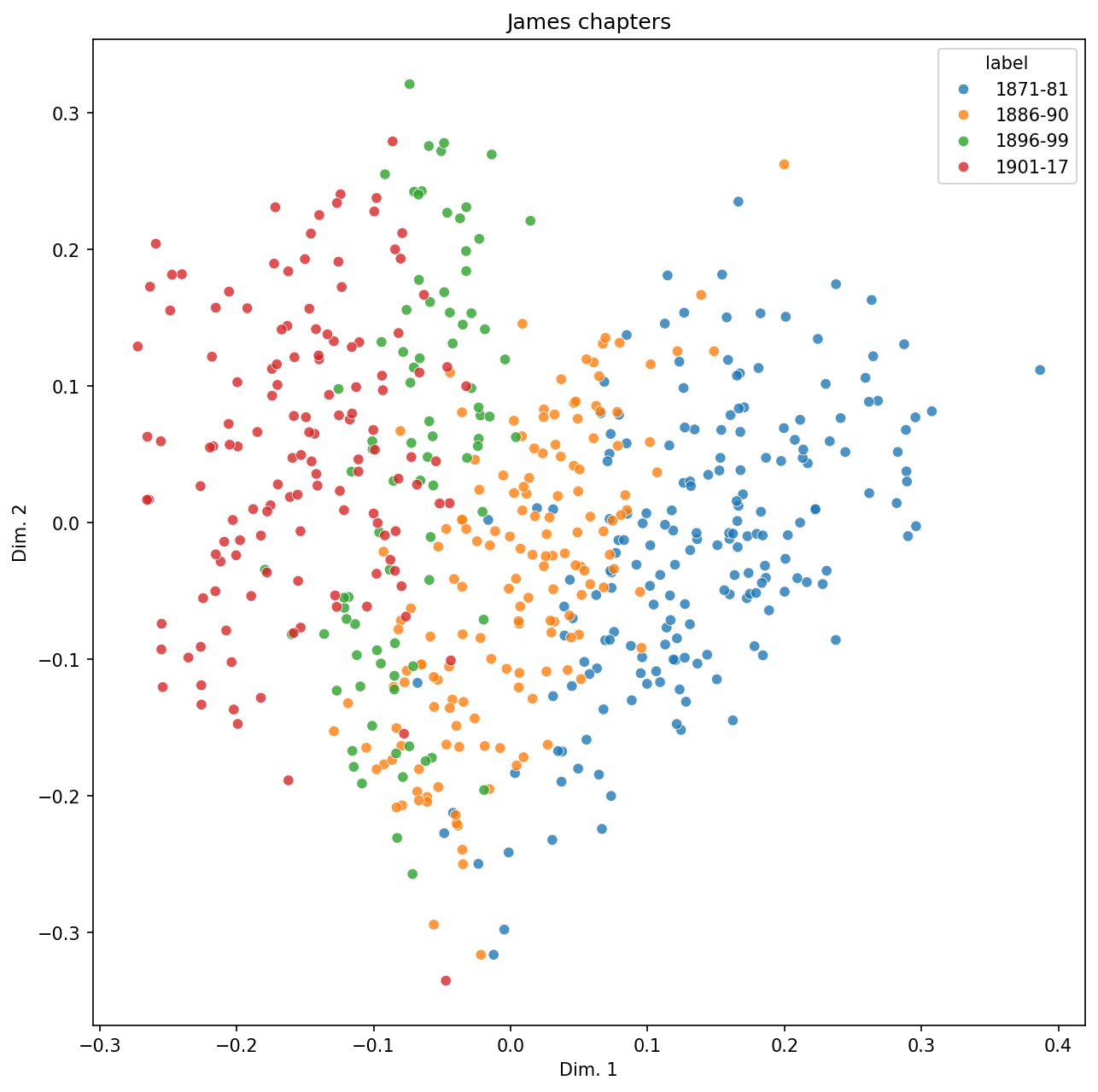
There isn’t perfect separation here. Might some of the overlapping points be mis-classified documents? We run predictions across all documents and re-plot with those.
all_preds = classifier.predict(tfidf)
Where are labels incorrect?
vis_data["incorrect"] = np.where(
all_preds == vis_data["label_idx"], False, True
)
Re-plot.
plt.figure(figsize = (10, 10))
g = sns.scatterplot(
x = "x",
y = "y",
hue = "label",
style = "incorrect",
size = "incorrect",
sizes = (300, 35),
palette = "tab10",
data = vis_data,
legend = "full"
)
g.set(title = "James chapters", xlabel = "Dim. 1", ylabel = "Dim. 2")
plt.show()
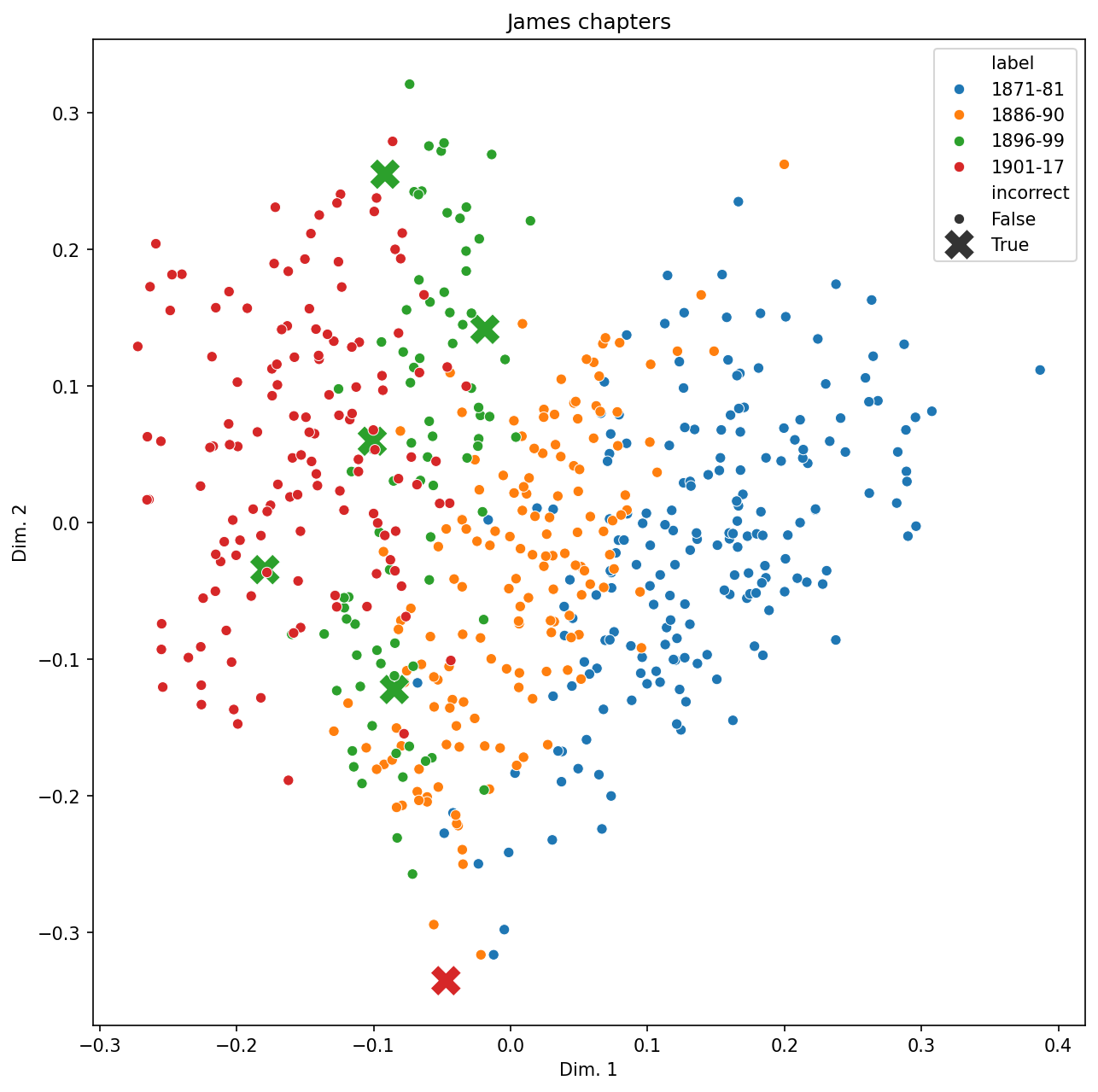
It does indeed seem to be the case that mis-classified documents sit right along the border of two classes. Though keep in mind that dimensionality reduction often results in visual distortions, so looking at data might sometimes be misleading.
Finally, which documents are these?
idx = vis_data[vis_data["incorrect"] == True].index
model_pred = all_preds[idx]
corpus.loc[idx, ["novel", "chapter", "hoover"]].assign(model_pred = model_pred)
| novel | chapter | hoover | model_pred | |
|---|---|---|---|---|
| 355 | Spoils Poynton | 13 | 2 | 3 |
| 384 | What Maisie Knew | 20 | 2 | 3 |
| 400 | Awkward Age | 5 | 2 | 3 |
| 422 | Awkward Age | 27 | 2 | 3 |
| 424 | Awkward Age | 29 | 2 | 3 |
| 562 | Ivory Tower | 13 | 3 | 1 |